

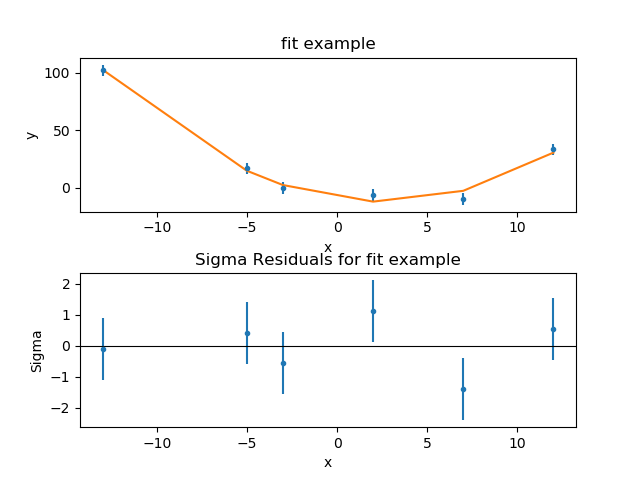
# code chunk number 15: grl-simul # set.seed( 1 ) P3 <- autoplot(ir, stat = "coverage", geom = "line", facets = sample ~. P2 <- autoplot(ir, aes(fill = pair)) + theme(legend.position = "none" ) # code chunk number 14: ir-exp # p1 <- autoplot(ir) # add meta data df <- DataFrame(value = rnorm(N, 10, 3 ), score = rnorm(N, 100, 30 ), Width = sample( 70 : 75, size = N,replace = TRUE )) Ir <- IRanges(start = sample( 1 : 300, size = N, replace = TRUE ), # code chunk number 13: ir-load # set.seed( 1 ) # code chunk number 12: seqinfo # autoplot(sq) # code chunk number 11: seqinfo-src # data(hg19Ideogram, package = "biovizBase" ) Layout_circle(gr, geom = "link", linked.to = "to.gr", radius = 6, trackWidth = 1 ) TrackWidth = 3, grid = TRUE, aes(y = score)) + Layout_circle(gr, geom = "point", color = "red", radius = 14 , Layout_circle(gr, geom = "bar", radius = 10, trackWidth = 4 , Ggplot() + layout_circle(gr, geom = "ideo", fill = "gray70", radius = 7, trackWidth = 3 ) +
# code chunk number 9: gr-autoplot-circle # autoplot(gr, layout = 'circle' ) # code chunk number 8: gr-facet-strand # autoplot(gr, stat = "coverage", geom = "area" ,įacets = strand ~ seqnames, aes(fill = strand)) Tracks( 'non-group' = p1, 'lfish = TRUE' = p2, 'lfish = FALSE' = p3) # lfish = FALSE, save space p3 <- autoplot(gra, aes(fill = group, group = group), geom = "alignment", lfish = FALSE ) p2 <- autoplot(gra, aes(fill = group, group = group), geom = "alignment" ) # in this way, group labels could be shown as y axis. # default is lfish = TRUE, each group keep one row. # when use group method, gaps only computed for grouped intervals. # if you desn't specify group, then group based on stepping levels, and gaps are computed without # considering extra group method p1 <- autoplot(gra, aes(fill = group), geom = "alignment" ) # code chunk number 6: autoplot.Rnw:236-237 # autoplot(gr, geom = "arch", aes(color = value), facets = sample ~ seqnames) # use value to fill the bar p2 <- autoplot(gr.b, geom = "bar", aes(fill = value)) # code chunk number 5: bar-default # p1 <- autoplot(gr.b, geom = "bar" ) Width = sample( 4 : 9, size = 10, replace = TRUE )), Gr.b <- GRanges(seqnames = "chr1", IRanges(start = seq( 1, 100, by = 10 ), # code chunk number 4: bar-default-pre # set.seed( 123 ) # code chunk number 3: default # autoplot(gr)
Idx <- sample( 1 : length (gr), size = 50 ) Sample = sample( c ( "Normal", "Tumor" ), Width = sample( 70 : 75, size = N,replace = TRUE )), Start = sample( 1 : 300, size = N, replace = TRUE ), , type = c("heatmap", "link", "pcp", "boxplot", "scatterplot.matrix"), ot = FALSE, # S3 method for class 'RangedSummarizedExperiment':Īutoplot(object. "t", rotate = FALSE, ot = FALSE, main_to_pheno "scatterplot.matrix", "pcp", "MA", "boxplot", Rownames.label = TRUE, colnames.label = TRUE,Īutoplot(object. Geom = NULL, type = c("viewSums", "viewMins", "viewMaxs", "viewMeans"))
, xlab, ylab, main, nbin = 30, binwidth,įacetByRow = TRUE, stat = c("bin", "identity", "slice"), Type = c("viewSums", "viewMins", "viewMaxs", "viewMeans"))Īutoplot(object. Geom = NULL, stat = c("bin", "identity", "slice"), , xlab, ylab, main, truncate.gaps =įALSE, truncate.fun = NULL, ratio = 0.0025,Ĭ("alignment"), stat = c("identity", "reduce"),Īutoplot(object. , xlab, ylab, main, which)Īutoplot(object, which. Resize.extra = 10, space.skip = 0.1, verage =Īutoplot(object. "estimate"), coord = c("linear", "genome"), , which, xlab, ylab, main,īsgenome, geom = "line", stat = "coverage", method = c("raw", , xlab, ylab, main, which,Īutoplot(object. Geom = NULL, stat = NULL, l = "gray50",Ĭoverage.fill = l, lfish = FALSE)Īutoplot(object. , xlab, ylab, main, indName = "grl_name", Layout = c("linear", "karyogram", "circle"))Īutoplot(object. Truncate.fun = NULL, ratio = 0.0025, space.skip = 0.1,Ĭoord = c("default", "genome", "truncate_gaps"), , chr, xlab, ylab, main, truncate.gaps = FALSE, Usage # S3 method for class 'GRanges':Īutoplot(object. Simpler and easy to produce fairly complicate graphics, though you may Genomic data compare to low level ggplot method, it is much Object, it tries to give better default graphics and customizedĬhoices for each data type, quick and convenient to explore your Autoplot: Generic autoplot function Description autoplot is a generic function to visualize various data


 0 kommentar(er)
0 kommentar(er)
